Group Clausen
Tim Clausen
Research Institute of Molecular Pathology (IMP), Campus Vienna Biocenter 1, 1030 Vienna, Austria
Phone: +43 (0)1 79730-3300, Email: tim.clausen(at)imp.ac.at
Molecular Mechanisms of Protein Quality Control
Our group studies complex proteases, chaperones and unfoldases that function as safeguards in the cell, preventing other proteins from undergoing dangerous interactions. In addition to these executive enzymes, we study regulatory factors, marking for example aberrant proteins that require special treatment. To elucidate the construction of the molecular machines implicated in protein quality control, we apply an integrative approach combining structural biology, protein biochemistry and molecular cell biology. By addressing the mechanistic details of protein folding and degradation pathways, we aim to better understand neurodegenerative diseases and to uncover strategies against mutations that effect protein integrity, as seen in cancer and ageing. Moreover, differences in the protease/chaperone systems between bacterial pathogens and their hosts will guide development of novel antibiotics.
Key Publications
* Hellerschmied, D., Roessler, M., Lehner, A., Gazda, L., Stejskal, K., Imre, R., Mechtler, K., Dammermann, A., Clausen, T. (2018) UFD-2 is an adaptor-assisted E3 ligase targeting unfolded proteins. Nat Commun. 9:484
* Trentini, DB., Suskiewicz, MJ., Heuck, A., Kurzbauer, R., Deszcz, L., Mechtler, K., Clausen, T. (2016). Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 539(7627):48-53
* Heuck, A., Schitter-Sollner, S., Suskiewicz, MJ., Kurzbauer, R., Kley, J., Schleiffer, A., Rombaut, P., Herzog, F., Clausen, T. (2016). Structural basis for the disaggregase activity and regulation of Hsp104. Elife. 5
Projects within VBC Ubiquitin Club
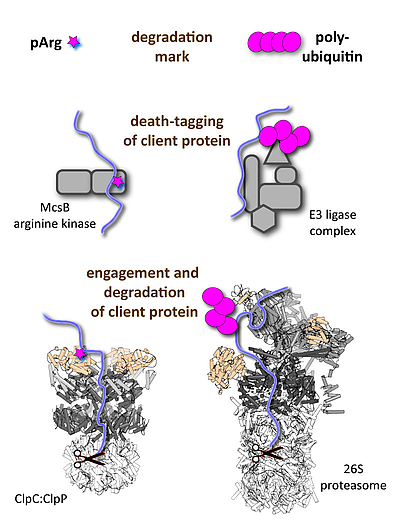
We are interested in studying “death-tagging” enzymes that mark specific client proteins to be degraded by dedicated protease complexes, such as the 26S proteasome in eukaryotes or the bacterial Clp proteases.
a) The Ub-Proteasome System: The ubiquitination of proteins is carried out by an enzymatic cascade comprising E1, E2 and E3 enzymes, of which the the E3 ligases is by far largest family attaching distinct Ub chains to cognate substrates. How the E3s select the right substrate at the right time and the right place and how they assemble Ub chains of varying types and lengths is a matter of intense research. We are currently characterizing various E3 ligases that are of high medical interest, being implicated in cancer related pathways or targeting disease relevant proteins. Moreover, our E3s of interest represent intricate molecular machines that are exciting on their own, having for example an extraordinary molecular size, complexity, substrate targeting mechanism or moonlighting function in the cell.
b) The pArg-ClpC:ClpP system: Energy-dependent proteases are essential for all living organisms to carry out protein quality control and degrade short-lived regulatory proteins. Despite employing distinct components, the eukaryotic and prokaryotic protease machines evolved a common bi-partite architecture. They are composed of a proteolytic core (20S proteasome, ClpP) for protein shredding and an associated AAA complex (19S regulatory particle, various Clp ATPases) that mediates substrate recognition, unfolding, and translocation. In contrast to eukaryotes, which universally employ poly-ubiquitin chains for marking client proteins, a general post-translational modification regulating proteolysis in bacteria was not known. A precious study from our lab characterized such a tagging system. It is the little-known modification phospho-arginine (pArg) that serves as degradation tag thus linking protein phosphorylation and protein degradation. Current projects address the substrate targeting mechanism of the “death-tagging” protein arginine kinase, McsB, the additional role of the pArg mark as functional switch, the distribution of protein arginine phosphorylation beyond Gram-positive bacteria, and how this unique degradation system could be hijacked to develop novel antibiotic drugs.